Geneticist Madhuri Hegde and her colleagues have a paper in the journal Genome Researchthat addresses the question: where do copy number variations come from?

Madhuri Hegde, PhD
Copy number variations (CNVs), which are deletions or duplications of small parts of the genome, have been the subject of genetic research for a long time. But only in the last few years has it become clear that copy number variations are where the action is for complex diseases such as autism and schizophrenia. Geneticists studying these diseases are shifting their focus from short, common mutations (often, single nucleotide polymorphisms or SNPs) to looking at rarer variants such as CNVs. A 2009 discussion of this trend with Steve Warren and Brad Pearce can be found here.
Hegde is the Scientific Director of the Department of Human Genetics’ clinical laboratory. Postdoctoral fellow Arun Ankala is the first author. In the new paper, Ankala and Hegde examine rearrangements in patients’ genomes that arose in 62 clinical cases of Duchenne’s muscular dystrophy and several other diseases. Mutations in the DMD gene are responsible for Duchenne’s muscular dystrophy.
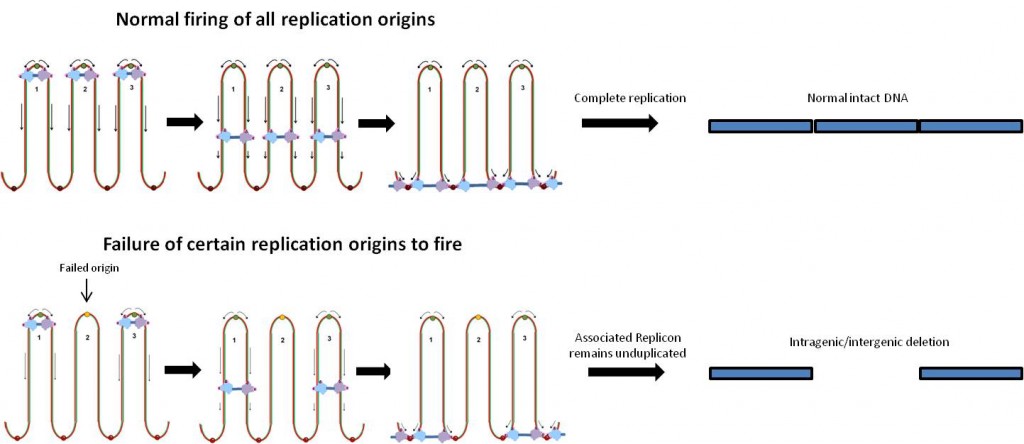
The pattern of the rearrangement hints what events took place in the cell beforehand, and hint that a problem took place during replication of the DNA. The signature is a tandem duplication of a short segment next to a large deletion, indicating how the DNA was repaired.
The authors note that the DMD locus is especially prone to these types of problems because it is much larger than other gene loci. The gene is actually the longest human gene known on the DNA level, covering 2.4 megabases (0.08 percent of the genome.)
Replication origins are where the DNA copying machinery in the cell starts unwinding and copying the DNA. Bacterial circular chromosomes have just one replication origin. In contrast, humans have thousands of replication origins spread across our chromosomes. In the discussion, the authors suggest that DNA copying problems may also explain duplications and historically embedded rearrangements of the genome.