Cancer cells have an array of built-in self-destruct buttons called death receptors. A drug that targets death receptors sounds like a promising concept, and death receptor-targeting drugs have been under development by several biotech companies. Unfortunately, so far results in clinical trials have been disappointing, because cancer cells appear to develop resistance pathways.

Death receptor-targeting drugs under development include: drozitumab, mapatumumab, lexatumumab, AMG655, dulanermin.
Winship Cancer Institute researcher Shi-Yong Sun, PhD and colleagues have a paper in Journal of Biological Chemistry that may help pick the tumors that are most likely to be vulnerable to death receptor-targeting drugs. This could help clinical researchers identify potential successes ahead of time and maximize chances of a good response for patients.
Postdoctoral fellow Youtake Oh is the first author. Winship deputy director Fadlo Khuri, MD and Taofeek Owonikoko, MD, PhD, co-chair of Winship’s clinical and translational research committee, are co-authors. Khuri’s 2010 presentation on death receptor drugs and lung cancer is available here (PDF).
Sun’s team shows that mutations in the cancer-driving genes Ras and B-Raf both induce cancer cells to make more of one of the death receptors (death receptor 5). In addition, they show that cancer cells with mutations in Ras or B-Raf tend to be more vulnerable to drugs that target death receptor 5.

Shi-Yong Sun, PhD
These mutations are known to be more common in some types of cancer. For example, roughly half of melanomas have mutations in B-Raf. Vemurafenib, a drug that inhibits mutated B-Raf, was approved in August 2011 for the treatment of melanoma. K-ras mutations are similarly abundant in lung cancer.
The selection and targeting of tumors via their specific mutations is a growing trend. Sun says lung, colon and pancreatic cancer are all cancer types where Ras and Raf mutations are common enough to become useful biomarkers. In lung cancer, Sun’s team’s results could be especially welcome news because, as a 2009 review concluded:
Recent studies indicate that patients with mutant KRAS tumors fail to benefit from adjuvant chemotherapy, and their disease does not respond to EGFR inhibitors. There is a dire need for therapies specifically for patients with KRAS mutant NSCLC.






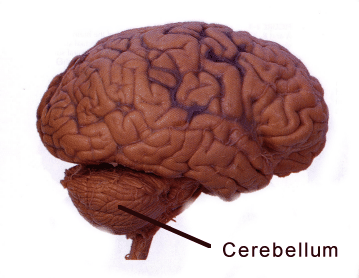 Hess and her colleagues discovered that drugs that stimulate AMPA receptors induce dystonia when introduced into the mouse cerebellum. Their results suggest that drugs that act in reverse, blocking AMPA receptors, could be used to treat dystonia.
Hess and her colleagues discovered that drugs that stimulate AMPA receptors induce dystonia when introduced into the mouse cerebellum. Their results suggest that drugs that act in reverse, blocking AMPA receptors, could be used to treat dystonia.