Move over, A, G, C and T. The alphabet of epigenetic DNA modifications keeps getting longer.
A year ago, we described research on previously unseen information in the genetic code using this metaphor:
Imagine reading an entire book, but then realizing that your glasses did not allow you to distinguish “g†from “q.†What details did you miss?
Geneticists faced a similar problem with the recent discovery of a “sixth nucleotide†in the DNA alphabet. Two modifications of cytosine, one of the four bases http://www.raybani.com/ that make up DNA, look almost the same but mean different things. But scientists lacked a way of reading DNA, letter by letter, and detecting precisely where these modifications are found in particular tissues or cell types.
Now, a team… has developed and tested a technique to accomplish this task.
Well, Emory geneticist Peng Jin and his collaborator Chuan He at the University of Chicago are at it again.
It turns out that 5-hydroxymethylcytosine or 5-hmC, the DNA modification Jin’s and He’s laboratories figured out how to read precisely, was just the beginning. Jin and He have a new paper in Cell describing a method for mapping a related modification: 5-formylcytosine. The co-first authors are U of C graduate student Chun-Xiao Song and Emory research specialist Keith Szulwach.
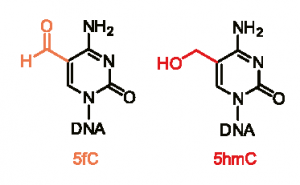 Using the “g†vs “q†analogy, it’s as if DNA has lots of g’s, some q’s, and a few letters that look similar to g but have an extra squiggle. What do those extra squiggles mean?
Using the “g†vs “q†analogy, it’s as if DNA has lots of g’s, some q’s, and a few letters that look similar to g but have an extra squiggle. What do those extra squiggles mean?
However, it gets complicated because DNA – the book of life – is subtly different in every cell, and is often being edited or repaired. The g’s and q’s aren’t simply static. Maybe those extra squiggles are editing marks that are going to get cleaned up soon? Or do they highlight words or Ray Ban outlet sentences that should be pronounced differently? That’s what Jin and He are trying to figure out.
Let’s back up a bit. The DNA modification generally known as methylation (5-mC) has been studied for decades. It is generally found on genes that are turned off, and it plays an important role in keeping them off. In contrast, 5-hmC – which is basically an oxidized form of 5-mC — appears to be enriched on active genes, especially in brain cells.
[Graduate student Tao Wang, in Steve Warren’s lab, has a recent paper on how the distribution of 5-hmC changes during human brain development.]
The focus of the new Cell paper, 5-formyl C, is an even more oxidized form of 5mC. Based on its distribution in the genomes of mouse embryonic stem cells, it looks like 5-formyl C is a sign that a cell is turning on a gene after it was turned off. 5-formyl C is significantly rarer, present at levels of 60 parts per million, compared to levels of a few percent for 5-hmC in some active genes.
Jin says: “5fC would represent active DNA demethylation. It is enriched at poised enhancers, but it is also present in other genomic features such as promoters or active enhancers.â€
A similar paper in Cell from Yi Zhang and colleagues at Harvard looks at the same issue. This is a particularly active area of research, since 5-hmC appears to have important functions in stem cells and the brain.

