Just a follow-up to last week’s announcement from the Emory Transgenic Mouse and Gene Targeting core that they are offering CRISPR/Cas9 gene editing for mice. Using CRISPR/Cas9 to produce genetically altered mice is a
substantial advance over the old way of doing knockouts and other manipulations (which itself won a Nobel Prize in 2007), mainly because it’s faster and easier.
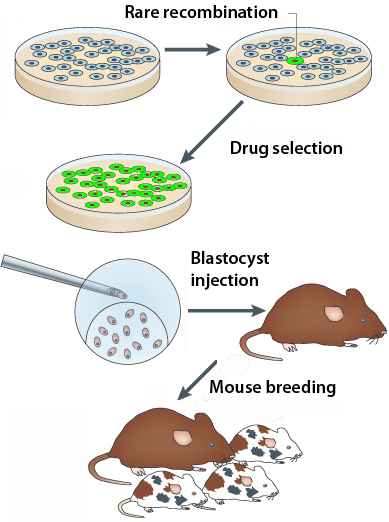
To appreciate the difference, consider that the old way involves introducing DNA into mouse embryonic stem cells, and then selecting for the rare cells that take up and incorporate the DNA in the right way. Then the ES cells have to be injected into a blastocyst, followed by mouse breeding to “go germline.”
With CRISPR/Cas9, it’s possible to inject pieces of RNA that target the desired genetic changes, straight into a one-cell stage mouse embryo. Not every embryo has all the right changes, but the frequency is high enough to inject and screen. As this review explains, it’s possible to introduce mutations into three genes at once and get mice quickly, rather than make each one separately and then breed the mice together, which can take many months.
Also, because of the need for drug selection, the targeting construct in old-school gene targeting has to be a blunt instrument. That can make it hard to make subtle changes to a gene — like introduce point mutations corresponding to natural variations linked with human disease — without taking a sledgehammer to the entire gene locus. CRISPR/Cas9 takes care of that problem.
Despite the advantages of this technology, three things to keep in mind:
*Many genetically altered mice are already available “off the shelf” as part of the International Knockout Mouse/Mouse Phenotyping Consortium.
*Emory’s Mouse Core has been working with the company Ingenious Gene Targeting, and has been out-sourcing some of the tedious aspects of old-school gene targeting in mice to Ingenious, starting last year. Technicians there can generate a dazzling array of conditional knockouts. If you want your favorite gene to flip around and produce a fluorescent protein when you give the mice an antibiotic, but only in some cells — Ingenious can do that. Old school is actually still the way to go for fancy stuff like this.
*Jackson Labs in Maine also works with Emory, offering similar services, and offers a guarantee. Read more