Emory biochemist Eric Ortlund participated in a study that was recently published in Proceedings of the National Academy of Sciences, which involves tinkering with billions of years of evolution by introducing mutations into DNA polymerase.
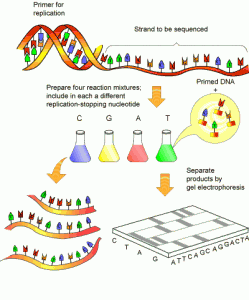
What may soon be old-fashioned: next-generation sequencing combines many reactions like the one depicted above into one pot
DNA polymerases, enzymes that replicate and repair DNA, assemble individual letters in the genetic code on a template. The PNAS paper describes efforts to modify Taq DNA polymerase to get it to accept “reversible terminators.” (Taq = Thermus aquaticus, a variety of bacteria that lives in hot springs and thus has heat-resistant enzymes, a useful property for DNA sequencing)
Ortlund was involved because he specializes in looking at how evolution shapes protein structure. Along with co-author Eric Gaucher, Ortlund is part of the Fundamental and Applied Molecular Evolution Center at Emory and the Georgia Institute of Technology.
To sequence DNA faster and more cheaply, scientists are trying to get DNA polymerases to accept new building blocks. This could facilitate next-generation sequencing technology that uses “reversible terminators” to sequence many DNA templates in parallel.




