Humans are good at deciphering complex images, compared to computers. Until recently, internet users often needed to verify that they were human by completing a CAPTCHA security check. A familiar variety asked the user to check all the boxes that contain a car, or a street sign.
If we asked random people off the street to look at pathology slides and “quick, check all the boxes that contain tumor cells,” what would happen? The accuracy, compared to a trained pathologist, wouldn’t be very good.
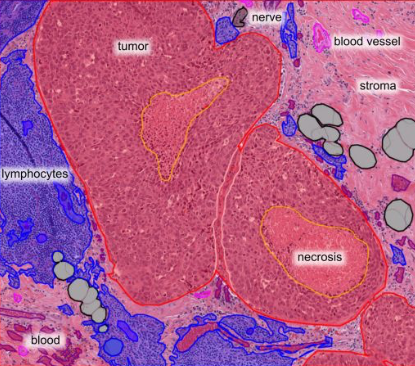
Not as easy as labeling which boxes contain street signs!
This challenge of expertise – crowdsourcing and pathology are not immediately compatible – is what Lee Cooper and colleagues sought to overcome in a recent paper published in Bioinformatics. So they put together something they called “structured crowdsourcing.”
“We are interested in describing how the immune system behaves in breast cancers, and so we built an artificial intelligence system to look at pathology slides and identify the tissue components,” Cooper says.
His group was particularly interested in the aggressive form of breast cancer: triple negative. They used pathology slide images from the Cancer Genome Atlas, a National Cancer Institute resource. The goal was to mark up the slides and label which sections contained tumor, stroma, white blood cells, dead cells etc.
They used social media to recruit 25 volunteers — medical students and pathologists from around the world (Egypt, Bangladesh, Saudi Arabia, United Arab Emirates, Syria, USA). Participants underwent training and used Slack to communicate and learn about how to classify images. They collaborated using the Digital Slide Archive, a tool developed at Emory. Read more




