Ribosomes, the factories that assemble proteins in cells, read three letters of messenger RNA at a time. Occasionally, the ribosome can bend its rules, and read either two or four nucleotides, altering how downstream information is read: frameshifting.
This week, Christine Dunham’s lab in the Department of Biochemistry has a paper in PNAS on how ribosomal frameshifting works, one of several she has published on this topic. The first author is postdoc Samuel Hong, now at MD Anderson. A commentary in PNAS calls their paper a “major advance” and “culmination of a half-century quest.”
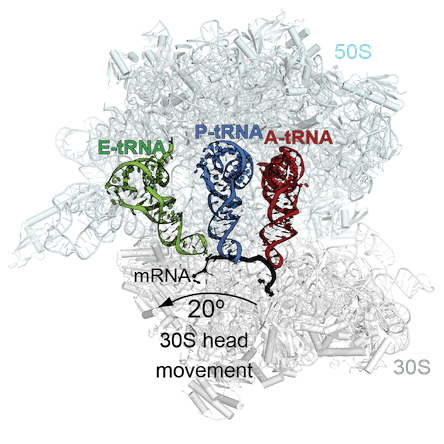
A suppressor tRNA can occupy more than one site on the ribosome. Adapted figure courtesy of Christine Dunham
Some antibiotics disrupt protein synthesis by encouraging frameshifting to occur, so a thorough understanding of frameshifting benefits antibiotic research. Also, scientists are aiming to use the process to customize proteins for industrial and pharmaceutical applications, by inserting amino acid building blocks not found in nature.
When mutations add or subtract a letter from a protein-coding gene, that usually turns the rest of the gene to nonsense. Compensatory mutations in the same gene can push the genetic letters back into the correct frame. However, others are separate, found within the machinery for translating the genetic code, namely transfer RNAs: the adaptors that bring amino acids into the ribosome. Suppressor tRNAs can compensate for a forward frameshift in another gene.
The Dunham lab’s new paper solves the structure of a bacterial ribosome undergoing “recoding” influenced by a suppressor tRNA. Her group had previously captured how the ribosomes decode this tRNA in one site of the ribosome, the aminoacyl or A site, in a 2014 PNAS paper. The new structures show how the tRNA moves through the ribosome out-of-frame to recode. The tRNA undergoes unusual rearrangements that cause the ribosome to lose its grip on the mRNA frame and allows the tRNA to form new interactions with the ribosome to shift into a new reading frame.

